Rapid Microbiological Methods for a New Generation
One of the greatest contributions to the field of microbiology came from the kitchen. In the 1880s, scientist Walter Hesse was searching for a solid medium that could be used to cultivate bacteria. The material had to be stable at high temperatures, and had to allow different types of microbes to be separated easily. Fanny Angelina, Hesse’s wife and laboratory assistant, had the answer: Agar-Agar, a gelling agent that she used in her jellies and puddings. This simple kitchen ingredient revolutionized the science of microbiology, allowing the separation and culturing of microbes to become a routine procedure.
The growth of microbial cells on agar surfaces may provide critical information about the number and the type of organisms that are present in a sample, but it can take days or even weeks to get results. In many cases, individual organisms cannot be isolated due to confluent growth, necessitating subculturing onto additional agar media, further delaying the time to result. In addition, many microbes may not replicate when cultured on artificial media after they’ve been deprived of nutrients or exposed to sublethal concentrations of preservatives, disinfectants, heat or decontaminating gases.
The modern microbiological laboratory needs more innovative approaches to microbial detection, identification and enumeration. Fortunately, technology is now available — or close to being available — that will speed up microbiological analysis, allowing aseptic manufacturing to embrace the concepts of Process Analytical Technology (PAT). This article surveys the various Rapid Microbiological Methods (RMMs) that are now available (or are in development) to the pharmaceutical industry.
PAT and FDA’s acceptance of rapid methods
The PAT initiative describes a regulatory framework that encourages the voluntary development and implementation of innovative approaches in pharmaceutical development, manufacturing and quality assurance (Pharmaceutical Manufacturing, November/December 2004, p. 41). Many new technologies are currently available that provide information on physical, chemical, and microbiological characteristics of materials to improve process understanding and to measure, control and/or predict product quality and performance. However, RMMs have not yet been widely implemented within the pharmaceutical industry due to concerns about validation and the need to prove the technologies’ “comparability” with existing methods, worries about return on investment (ROI) and regulatory uncertainty.
Many forward-looking microbiologists working at pharmaceutical companies are frustrated by their management’s hesitancy to approve the use of RMMs. Typically, these managers fear that the company won’t be able to make a compelling case to regulators that the new methods work as well or better than technologies that the company already uses, or that there’s not enough guidance available to validate a new method and/or laboratory system. This simply is not true. Companies that have conducted successful validations or those that are now qualifying new systems have relied on guidelines presented in PDA Technical Report #33, “Evaluation, Validation and Implementation of New Microbiological Testing Methods.” Published in 2000, this document provides information on validation protocol design, testing and acceptance criteria, and installation, operational and performance qualification strategies.
But demonstrating comparability of the new technology is only one part of the overall strategy for embracing RMMs. Management may also be concerned that:
- Technologies may be incompatible with product and/or processes;
- Staff may not understand what the required method sensitivity or specificity should be (e.g., detection levels and for what types of microorganisms);
- Technologies may require significant operator training and qualification;
- Technology vendors won’t be able to provide support during the initial assessment, validation exercises, and most importantly, after the system has been placed in service for routine use.
Due diligence is essential, from both a technical and a business perspective, when implementing RMMs, to ensure that any technology selected will be the best possible fit for the application involved.
Making a business case for RMMs
RMMs are not inexpensive. The overhead alone can run into the hundreds of thousands of U.S. dollars, required for:
- initial feasibility assessment;
- development of user requirements;
- testing protocols and SOPs;
- training;
- execution of the validation program;
- documentation;
- regulatory submission;
- technology transfer;
- site implementation.
These expenses don’t include the costs for the equipment itself, or testing costs, which tend to be higher than those for established technologies.
At any pharmaceutical company, corporate management will demand to know what ROI it can expect from these new technologies. In order to provide the most accurate information, any team exploring the use of RMMs must include someone from the company’s financial or business planning department. Their expertise will ensure that the budget is accurate, and that the team makes a strong business case for the technology. In many cases, the long-term benefits of RMMs will far outweigh the short-term costs.
Technical requirements for any RMM might include the following:
- Significantly reduced time-to-result when compared with conventional microbiological methods;
- Automated, miniaturized and high-throughput technology platforms;
- Increased sensitivity, accuracy, precision and reproducibility;
- Detection of a single, viable microorganism without the requirement for cellular growth;
- Enhanced detection of stressed organisms.
Business requirements might include:
- Significant reduction of testing time to release products more rapidly;
- Lower inventories (raw material, in-process material and finished product;
- Prevention of back orders;
- Reduction of repeat testing, deviations, OOS investigations and product rejection.
Regulatory uncertainty
Regulatory uncertainty is impeding the drug industry’s acceptance of RMMs and other new technologies today. Many drug companies view the existing regulatory system as rigid, and fear that FDA inspectors may not be familiar with recent advances in microbiology.
Quality and manufacturing professionals may have legitimate concerns or questions, ranging from fear of increased sensitivity in recovered microbial counts to concerns about the potential impact on existing acceptance criteria, uncertainties over the development of a meaningful validation and submission strategy, or doubts that a particular technology platform may be acceptable for use with a given product or process. Teams evaluating RMMs should be proactive, contact FDA, meet with the Agency’s microbiologists and discuss the best path forward. The spirit of FDA’s PAT Guidance views such communication as essential to advancing drug manufacturing technologies.
The U.S. and European Pharmacopeias (USP and EP) have already proposed general chapters on rapid microbiological methods. The planned USP informational chapter <1223>, “Validation of Alternative Microbiological Methods,” provides guidance for validating methods that can be used as alternatives to official Compendial microbiological methods. The proposed informational EP chapter 5.1.6, “Alternative Methods for Control of Microbiological Quality,” describes alternative methods for the control of microbiological quality.
Applications and opportunities
There are many opportunities for implementing RMMs in the pharmaceutical industry, including:
- Raw material and component testing;
- in-process and pre-sterilization/filtration bioburden;
- fermentation and cell culture monitoring;
- purified/process water testing;
- environmental monitoring (e.g., surface, air, compressed gases, personnel);
- bacterial endotoxin testing;
- microbial limits;
- antimicrobial effectiveness testing;
- biological indicator survival studies;
- sterility testing;
- media fill failure investigation;
- contamination incident assessment.
RMMs can be applied as PAT, when information about the microbial control of a manufacturing process can be obtained in real-time — for example, in purified water testing, in-process bioburden testing and environmental monitoring. In such cases, a company can apply RMMs to respond, immediately, to an OOS finding or an adverse trend, minimizing the impact to product or in-process material. The alternative is to wait for days until conventional testing results are available, after the opportunity to act has long passed.
Many RMMs offer increased sensitivity, accuracy, precision and reproducibility, and some can detect a single cell without the need for microbial growth. As more rapid method technology platforms are developed, instruments are being miniaturized to facilitate at-line, on-line and in-line applications at the site of manufacturing. RMMs can enhance risk-based analysis and microbial control strategies by offering a more robust understanding of manufacturing processes.
The following section provides an overview of the RMM platforms available or being developed, with a brief discussion of how they work.
Growth-based technologies rely on the measurement of biochemical or physiological parameters that reflect the growth of microorganisms. These types of systems require the organisms in a sample to proliferate, either on a solid or liquid medium, in order to be detected and/or quantified. Currently available growth-based technologies for the detection, enumeration and identification of microorganisms include:
- the bioMérieux (Durham, N.C.) VITEK 2 and Bactometer systems;
- the Biolog (Hayward, Calif.) Omnilog
- ATP-bioluminescence systems such as Millipore’s (Billerica, Mass.) Milliflex Rapid System, the Celsis (Chicago) Advance Luminometer, and Pall’s (East Hills, N.Y.) PallChek Microbiology System;
- Genomic Profiling Systems’ (Bedford, Mass.) Growth Direct, a new system, currently in development, that relies on the growth of microorganisms on agar.
Viability-based technologies use viability stains and/or cellular markers to detect and quantify microorganisms without the need for cellular growth. Today’s available technologies include:
- the Chemunex (Princeton, N.J.) ScanRDI solid-phase cytometry platform;
- flow-cytometry systems such as the Chemunex D-Count and BactiFlow, and the Advanced Analytical Technologies (Ames, Iowa) RBD 3000.
Artifact-based technologies rely on the analysis of cellular components or the use of probes that are specific for microbial target sites of interest. Examples include:
- the MIDI (Newark, Del.) Sherlock Microbial Identification System;
- the Waters (Milford, Mass.) MicrobeLynx system utilizing MALDI time-of-flight mass spectrometry;
- SELDI time-of-flight mass spectrometry using Ciphergen (Fremont, Calif.) ProteinChip Arrays;
- a portable, hand-held assay platform for the detection of endotoxin and Gram-negative bacteria using the Charles River Laboratories (Wilmington, Mass.) Endosafe PTS system;
- the Cambrex (East Rutherford, N.J.) PyroSense on-line endotoxin detection system, which is currently in development, and is intended to monitor purified water systems.
Nucleic acid-based technologies rely on PCR DNA amplification, 16S rRNA typing, gene sequencing and other novel applications. Many of these systems are used for the rapid and accurate detection of a specific target microorganism or for the identification of an unknown isolate. Although these systems can provide results much faster than the growth-based detection and identification technologies, they still require starting material from a microbial culture (e.g., an isolated colony on an agar surface), and therefore will add additional time in obtaining the final result. Commercially available technologies that utilize nucleic acid-based technologies include:
- the DuPont Qualicon (Wilmington, Del.) RiboPrinter and BAX systems;
- Applied Biosystems’ (Foster City, Calif.) MicroSeq;
- Sequenom’s (San Diego, Calif.) MassARRAY platform;
- Bacterial BarCodes’ (Houston) DiversiLab System;
- the Ibis (Carlsbad, Calif.) PCR-mass spectrometry TIGER system.
On the cutting edge of miniaturization
Other technology platforms are much smaller than the technologies previously discussed, and should have a significant impact in the way microbiological assays will be performed in the future. Some of these platforms offer continuous and instantaneous microbial detection, and are excellent candidates for at-line, on-line or in-line microbiological PAT applications. The technologies include biochips, microarrays, biosensors, and an instantaneous detection system for airborne microorganisms.
Micro-Electro-Mechanical Systems (MEMS)
Imagine, for a moment, a machine so small that the human eye cannot see it and thousands of these machines are manufactured on a single piece of silicon. Imagine a future where gravity and inertia are no longer important, but atomic forces and surface sciences dominate. This is the world of MEMS, and the future is now.
MEMS integrate mechanical, electrical, fluidic and optical elements, sensors, and actuators on a common silicon substrate, using microfabrication technology. Use of MEMS is growing the fastest in drug discovery and delivery. However, many of the same technologies used in these applications already have, or soon will have, a place as RMMs. They include the following platforms:
Lab-On-A-Chip is based on an automated, micro- or nano-scale laboratory that enables sample preparation, fluid handling and analysis and detection steps to be carried out within the confines of a single microchip. The technology is based on microfluidics, and the most familiar consumer application is ink-jet printing. Microfluidics allows for the manipulation of minute amounts of liquid in miniaturized systems that are composed of a network of channels and wells that are etched onto glass or polymer chips. Pressure or voltage gradients move pico- or nanoliter volumes through the channels in a finely controlled manner that enables sample handling, mixing, dilution, electrophoresis and chromatographic separation, staining and detection. Currently available lab chips analyze protein, DNA, RNA and whole cells in fluid samples. Examples that are now available include:
- Bacterial Barcodes’ DiversiLab Microbial Typing System, which uses a microfluidics chip (the DNA LabChip) to separate rep-PCR amplicons. The chip is processed in a bioanalyzer, where the amplicons pass through a laser, causing fluorescence of an intercalating dye. The resulting rep-PCR fingerprints are compared with a database, and a detection result, or microbial identification, is provided.
- A micro-scale impedance-based detection system is currently being developed by BioVitesse (San Francisco, Calif.) and Purdue University. In impedance microbiology-based systems, bacterial growth is detected by monitoring the movement of ions between two electrodes (conductance) or the storage of charge at the electrode surface (capacitance). Conventional systems require 104 - 105 cells in milliliter-sized samples in order to elicit a positive response over time. However, if the number of cells can be concentrated into a small incubation chamber, the time to detect microbial growth should decrease. This is the basis for the BioVitesse system. On a single chip, sample channels and incubation chambers are etched onto a crystalline silicon substrate. Platinum microelectrodes, which are located in the incubation chambers, measure impedance changes. Because the sample volume in this system is less than 1 nanoliter, the time to detect microbial growth may be considerably less than what is currently available today.
Microarrays (Biochips or DNA Chips)
Microarrays are collections of miniaturized test sites, arranged on solid substrates, that permit many tests to be performed at the same time. They are composed of an orderly arrangement of protein or thousands of DNA or RNA fragments on glass, silicon or nylon substrates. This technology evolved from Southern Blotting technology, in which fragmented DNA is attached to a substrate and then probed with a known gene or DNA fragment, using fluorescent tags to allow visual detection.
Microarrays are usually fabricated using a variety of technologies, including printing with fine-pointed pins onto glass slides, photolithography, ink-jet printing or electrochemistry. Other methods may also be used, such as in situ synthesis, whereby the probes are synthesized directly on the chip instead of spotting them on the array. An example would be CombiMatrix’s (Mukilteo, Wash.) CustomArray.
Applications for microarrays include nucleic acid sequence identification and measuring expression levels of genes. For example, Affymetrix’s (Santa Clara, Calif.) GeneChip contains the entire human genome (~50,000 known genes and gene variants) on a single chip, while CombiMatrix’s CustomArray contains probes that can detect influenza A and Avian H5N1 (bird flu) strains. Both of these platforms are commercially available.
Biosensors
A biosensor can detect an analyte that is comprised of a biological component combined with a physicochemical detector component. Nineteenth-century miners used a canary in a cage as a biosensor to detect lethal concentrations of gas. Today, the most widespread example of a commercially available biosensor is the blood glucose monitor, but future uses will include remote sensing of airborne bacteria and detection of pathogens.
The biological component of a biosensor may contain tissue, microorganisms, organelles, cell receptors, enzymes, antibodies, nucleic acids or whole cells. The detector element works in a physicochemical manner and may include optical, electrochemical, thermometric, piezoelectric or magnetic technologies. An example of a currently available biosensor that performs immunological assays is the ICS Chip by Ambri (Chatswood, NSW, Australia). In this platform, the immunoassay components are spotted in 1-micron tall wells using less than 1 nanoliter of material.
Combination systems
Combining multiple MEMS platforms may result in a hybrid system that is superior to the individual platforms themselves. An example is ST Microelectronics’ (Geneva, Switzerland) In-Check silicon chip, which combines microfluidics and microarrays. This technology platform provides a fully integrated PCR reactor and a microarray used to hybridize and detect the PCR amplicons. The chip is mounted on a one-by-three-inch plastic slide that provides the necessary mechanical, thermal, electrical and fluidic connections.
A sample size of 2-8 µl is used, PCR reactions are three times faster than conventional thermocyclers, and a portable, customized fluorescent-based optical reader analyzes the microarray in a few seconds. The currently available platform hosts a pathogen panel to identify 10 sepsis-causing bacterial species as well as methicillin-resistant strains of Staphylococcus aureus from positive blood culture samples.
Nanotechnology
Nanotechnology operates at the atomic, molecular or macromolecular range of approximately 1 to 100 nanometers to create and use structures, devices, and systems that have novel properties. The key to this technology is high-voltage electron-beam (or E-beam) lithography, in which a beam of electrons is scanned across a surface covered with a thin film, called a resist. The electrons produce a chemical change in the resist, which allows the surface to be patterned.
Nanoarrays are the next evolutionary step in the miniaturization of bioaffinity tests for proteins, nucleic acids and receptor-ligand pairs. These arrays utilize approximately 1/10,000th of the surface area occupied by a conventional microarray, and over 1,500 nanoarray spots would occupy the area required for a single microarray spot.
For example, BioForce Nanosciences’ (Ames, Iowa) NanoArray prints biological and non-biological materials onto silicon chips (at right) and other surfaces with ultra-micro spot sizes ranging from 1-20 µm, and in the nanometer range to 250 nm. This technology utilizes surface patterning tools, or SPTs, microcantilever-based micro-fluidic handling devices. The droplet volumes these microcantilevers deliver to the nanoarray are in the femtoliter and attoliter range (10-12 milliliter and 10-15 milliliter, respectively).
Nanoarrays offer a number of advantages, including label-free detection (via atomic force microscopy). They work in both solutions and biological liquids, retaining biological activity for subsequent analyses.
Optical spectroscopy and instantaneous detection
Optical spectroscopy measures the interactions between light and the material being studied. Light scattering is a phenomenon in which the propagation of light is disturbed by its interaction with particles.
In a Mie scattering particle detector, airborne particles intersect a light beam emanating from a laser diode. If the air is free of particles, a “beam blocker” at the center of the first convex lens stops this laser beam. If the air contains particles, they will scatter the laser beam and cause part of the light to deviate at an angle from the incident beam. This scattered light will be collected by the second lens and focused onto a photo detector, which converts the light intensity to an electrical signal. This is the basis for a novel, instantaneous and continuous microbial detector, currently in development at BioVigilant Systems (Tucson, Ariz.).
The technology was originally developed to detect potential bioterrorism agents such as anthrax spores, but is now being adapted to the pharmaceutical industry’s unique needs. The platform uses a Mie scattering particle counter with effective detection of particles within the 0.5 to 20 µm range. The detection of biological particles is based on the autofluorescence of cellular targets including NADH and riboflavin, under UV laser with a wavelength of 405 nm. The company will be working on a method to capture microorganisms after analysis for further processing, such as microbial identification.
In short, these are very exciting times for microbiology, and, as more alternatives become available, more pharmaceutical microbiologists will embrace rapid detection, quantification and characterization technologies. No doubt Fanny would be proud of these worthy successors to agar cultivation.
About the Author
Michael Miller, Ph.D., is Senior Research Fellow in the Manufacturing Science and Technology (MS&T) function at Eli Lilly and Co. (Indianapolis). He is responsible for providing technical leadership in microbiology and sterility assurance within Manufacturing, Quality, Engineering, and Product Development. He is also accountable for leading Lilly’s corporate initiatives for Process Analytical Technology (PAT), barrier isolation technology and rapid microbiological methods. Previously, he held numerous business development, Quality and R&D leadership roles at Bausch & Lomb and Johnson & Johnson.
Dr. Miller has authored over 60 technical publications and presentations in the areas of rapid microbiological methods, PAT, ophthalmics, disinfection and sterilization, and has served as Chairperson for numerous rapid microbiological methods technical conferences in the United States and Europe. Most recently, he was the editor of the Encyclopedia of Rapid Microbiological Methods, a three-volume reference co-published by the PDA and Davis Healthcare International Publishing.
Dr. Miller holds a Ph.D. in Microbiology and Biochemistry from Georgia State University (GSU), a B.A. in Anthropology and Sociology from Hobart College, and has served as an adjunct professor at GSU and the University of Waterloo School of Optometry.
GSK: Using Rapid Detection for PAT-worthy Product Release
A microbial monitoring milestone was achieved in 2004 when GlaxoSmithKline announced that it had been granted FDA approval to use ATP bioluminescence technology to release a prescription nasal spray product in its Parma, Italy plant. This was the first time a drug maker had used a rapid detection technology to release product under FDA’s Process Analytical Technology (PAT) initiative.
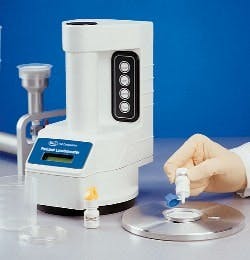
GSK Parma uses a novel two-tier release strategy for the non-sterile product: first, a presence/absence test using Pall Life Sciences’ (East Hills, N.Y.) Pallchek ATP bioluminescence system (at right); then, if contamination is present, further conventional assessment to determine its level and identity. The approach has allowed GSK to reduce its release time for the product from several days to 24 hours, said Newby. The approval was noteworthy for the fact that FDA had previously stated publicly that it would not accept such a strategy, Newby noted.
ATP bioluminescence is also being tested by Bristol-Myers Squibb in France to obtain immediate readings for monitoring Water-for-Injection system contamination, Dr. Eric Bagur, bacteriological quality control manager of BMS, France, recently reported. Several years ago, GSK gained European approval to use another rapid method, solid-phase cytometry, using the Chemscan RDI by Chemunex (Princeton, N.J.), for rapid testing of pharmaceutical-grade water.