Making antibody medicines manufacturing a reality
Antibody medicines are the most typical biologic drug and mainly consist of protein produced by genetically modified animal cells. The process of manufacturing antibody medicines is extremely complex and costly because the cell culturing process requires prolonged incubation times, along with a high level of sterility for handling living cells.
Complex biological processes involving cells are used for manufacturing these medicines, with new technologies continuously being developed to improve the quality and yield of these products. A study was conducted to clarify the relation between cell conditions and the yield of target products, and to investigate measurement methods required for improved control.
This article describes the technologies necessary for achieving these goals and reports the results of experiments conducted to verify effectiveness.
Challenges in cell culturing
When manufacturing antibody medicines, the yield of the target product is determined by the rate of production by each cell and the cumulative cell density during the culture period. Productivity depends largely on the culture medium (the solution in which the cells are cultured) and maintaining a proper growth environment suitable for high-density cell culture.
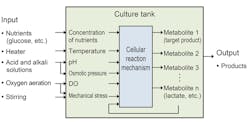
In the culture process, various parameters are measured and controlled to maintain appropriate conditions for cell growth. Figure 1 shows a typical culture tank used for the culture process and its inputs and outputs .1 Among the parameters, temperature, pH and the dissolved oxygen (DO) concentration are the primary targets for inline measurement and control because these parameters fluctuate quickly during the process.
In contrast, other parameters, such as the concentration of nutrient components, metabolic products, and target products, along with osmotic pressure, are not suited for inline measurement and hence have traditionally been handled by sampling and offline analyses. However, the frequency of such sampling and analyses is usually limited to once a day because sampling reduces the number of cells and the amount of the culture solution. Therefore, deviating culture conditions cannot be monitored in real time and appropriate actions cannot be taken quickly. Furthermore, sampling also increases the risk of contamination.
Since glucose is a major nutrient component for cells, it is essential for culture systems to maintain an ideal concentration. Conventionally, this concentration is controlled based on data collected from daily offline analysis. Depending on the analytical results, the glucose solution estimated to be consumed over the next several days is fed into the process manually. Therefore, the actual concentration can vary greatly, sometimes as much as 1 to10 g/L during the culture period, and sharp rises in glucose concentration are known to negatively affect growth conditions for many cell types.
It is well known that an overdose of glucose causes excessive accumulation of toxic metabolic products such as lactate and ammonia. Three steps are required to improve glucose measurement and control:
- Measure glucose concentration, lactate concentration and cell density inline
- Predict cell conditions (metabolism and growth) online based on these measurements to predict glucose consumption
- Maintain glucose concentration within a target range by automatically feeding glucose solution based on predicted glucose consumption
Defining the problem
Glucose and lactate concentrations can be measured inline by using near-infrared spectrometry to quantify concentrations based on light absorption linked to molecular vibration. By immersing a sensor probe directly in the culture medium, this method can be used to simultaneously measure both chemical compounds.
Cell density is measured by using impedance spectroscopy to quantify the number of living cells based on their electrical characteristics. Inline measurement can also be performed by immersing a sensor probe directly in the culture medium.
Regarding step two, cellular metabolism can be determined based on the glucose consumption rate (rate of consumption of glucose per cell) and the lactate production rate (rate of production of lactate per cell). Similarly, cellular growth can be determined based on the specific growth rate (rate of growth per cell).
The glucose consumption rate is calculated based on changes in glucose concentration and cell density, and the lactate production rate is calculated in a similar manner. The growth rate is calculated based on changes in cell density. Thus, all three rates needed to indicate and predict ideal cell conditions can be made by inline measurements.
Glucose consumption per unit volume can be determined by the glucose consumption per cell and the number of cells. These two factors can be obtained from inline measurement. When the volume of the culture solution is known, total glucose consumption can be predicted.
In practice, cell counts fluctuate during the culture process, and glucose consumption rate per cell also varies, causing large fluctuations in the total glucose consumption during a single run. Online prediction of glucose consumption enables glucose concentration to be controlled depending on culture conditions, as described in detail later.
With regard to step three, to maintain the glucose concentration within the target range, glucose solution is automatically fed to the culture medium based on the difference between the current glucose concentration and the target value, and on the current glucose consumption. Observations indicated it took some time before the effect of the glucose solution fed to the culture medium was reflected in the measurements. This issue was addressed by formulating an equation to optimize the feed rate of glucose solution to maintain the glucose concentration within the specified range (for example, 2 g/L ± 50%).
Inline sensors for measuring glucose concentration, lactate concentration and cell density are described below. Subsequently, a method of predicting cell conditions and controlling glucose concentration based on the prediction results is also described.
Measuring glucose and lactate concentrations
The enzyme electrode method is one popular choice for in-line concentration measurements. It detects a target component by using enzyme reactions and is widely used for measuring nutrient and metabolic components, such as glucose and lactate, in culture solutions. However, this method presents problems: enzymes tend to lose activity and thus the electrodes become unstable, and enzymes are often dissipated into solutions. Therefore, it is not recommended to immerse an enzyme electrode sensor directly in a culture tank.
Optical sensors avoid these issues and are thus better suited for inline measurement. In addition, optical sensors can measure multiple components simultaneously. In this application, Yokogawa’s InfraSpec NR801JL near-infrared analyzer was used for inline measurement of glucose and lactate.
Three factors must be considered when measuring components in culture solutions using an inline optical sensor: (a) effect of light scattered by cells, (b) extremely low concentration of the target components, and (c) existence of multiple components with a similar molecular structure.
Regarding point (a), preliminary experiments showed that the transmitted light intensity decreases to 1/10 as culturing advances from the initial stage with a low cell density to the middle stage with a high cell density. Regarding point (b), optical absorption by the target components is extremely low because more than 99 percent (mole fraction) of a culture solution is water, and the amount of the target components for measurement (glucose and lactate) is less than 0.1 percent. A sensor with a high signal-to-noise (S/N) ratio is required to address these two issues.
Therefore, the S/N ratio of the analyzer was increased by replacing its optical fiber with a 400 μm core diameter to one with a 600 μm core to reduce the coupling loss between the optical fiber and the sensor probe, and to increase the amount of light received by the sensor.
Regarding point (c), it is unavoidable that measurements are affected by components with a similar molecular structure because near-infrared spectrometry calculates the concentration of components based on light absorption caused by molecular vibration. In general, absorption spectra in the near-infrared region are a complex superposition of multiple broad peaks.
Therefore, the concentration of the target component is calculated based on absorption data (absorption spectra) at multiple wavenumbers instead of a single wavenumber. The equation used for converting the absorption spectra into the concentration of the target component is called a calibration curve. To minimize the effects of component with molecular structures similar to that of the target component, it is necessary to create a calibration curve that is highly selective and responsive to the target component.
Calibration curves are prepared by using culture solutions containing cells or without cells. For this application, a culture solution without cells was used to avoid light scattering by cells and ensure a high signal to noise ratio. This enabled the creation of calibration curves with high selectivity based on the absorption data of the target component.
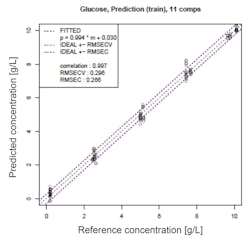
Exhibit 2 shows the relationship between glucose concentrations measured by a reference analyzer and the inline sensor.
Both measurements are in close agreement, with a root mean square error of cross validation (RMSECV) of 0.3 g/L, which is satisfactory for practical use. The results of inline measurement during the cell culture process are described later.
Measuring viable cell density
Trypan blue staining is a common method for measuring the density of living cells in the culture. The viable cell density is obtained by counting the number of dead cells stained by trypan blue dye, which requires sampling.
In contrast, impedance spectroscopy can measure viable cell density inline. Since a living cell is composed of electrically conducting cytoplasm and non-conducting cell membrane that wraps the cytoplasm, it can be regarded as a capacitor. Electric polarization does not occur in dead cells due to damage to cell membranes and other causes. Therefore, the capacitance component obtained by measuring the impedance of a culture solution is proportional to the overall living biomass, which can be converted to cell density data.
In the late stage of the culture process, however, the cell density reaches a peak after which the number of dead cells starts to increase, and the capacitance component is no longer proportional to the number of living cells. A possible reason is that as cell dimensions and membrane characteristics change during the culture process, the capacitance of each cell diverges from the initial stage.
However, viable cell density can be measured with high precision throughout the culture period by identifying the transition point of the relationship between the capacitance and the number of living cells, and by using different calibration curves before and after this point.
A method of detecting the transition point was developed by using capacitance data at multiple frequencies based on the impedance spectroscopy, and by automatically switching over among different calibration curves. This method enabled an appropriate calibration curve to be selected for each culture phase, improving measurement precision.
Exhibit 3 shows measurement results obtained by the trypan blue staining method, conventional impedance spectroscopy and the method described above.
The difference between the trypan blue staining method and the measurement results of the new method remained within 20 percent, sufficient for this application.
Predicting cell conditions
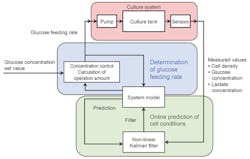
A method for predicting cell conditions online using measurement values from the inline sensor described above was developed, along with a method for predicting glucose consumption in a culture vessel based on predicted cell conditions. These factors are used for controlling the glucose feed rate as depicted in Exhibit 4.
An idea was formulated for predicting cell conditions based on measurement data from the inline sensors. A non-linear Kalman filter was used to reduce signal noise. Equations were used to express cell density, glucose concentration, lactate concentration and the time evolution of each specific rate. An equation was developed to calculate the rate of change of glucose concentration as the difference between the increment due to the feeding of the glucose solution and the decrement due to the consumption by cells.
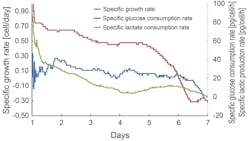
This model was repeatedly modified to make predicted values closer to sensor measurement values. To confirm the practicability of this online prediction model, the model was applied to a Chinese hamster ovary (CHO) cell culture system. As shown in Exhibit 5, this method produced satisfactory results in online prediction of changes in cell conditions (specific growth rate, specific glucose consumption rate, and specific lactate production rate).
Controlling glucose solution feed rate
Glucose consumption rate varies depending on cell conditions. Based on the online prediction of cell conditions, the glucose solution feed rate can be controlled as shown in the blue area of Exhibit 4.
The total glucose consumption in a culture vessel is calculated from the predicted cell density and the actual glucose consumption rate, along with the actual volume of culture solution. By combining the conditions above with other parameters such as pump speed, an equation was derived to optimize the glucose solution feed rate, and a system was developed to solve this equation online in a sequential manner. This system can automatically feed glucose solution to maintain the glucose concentration within a specified range, for example, 2 g/L ± 50%.
Developing an integrated solution
With each of the technologies described above now proven, the building blocks are in place to develop an integrated solution for biomedical production. Yokogawa has been developing a solution that integrates a culture system, inline sensors and control unit to regulate culture solution based on predicted cell conditions. Repeatable, satisfactory results have been obtained due to the high measurement precision of each inline sensor, which enables the successful prediction of cell conditions and the control of glucose concentration by the advanced control function. Based on these promising results, Yokogawa believes the automated manufacturing of antibody medicines will become a reality in the near future.