Leveraging AI to Drive a Virtuous Cycle Between Drug Development and Application
Advances in machine learning and artificial intelligence (AI) are transforming insight across a variety of scientific fields and industries. In this article, we explore the power of these advances to change the way information and analysis are used in drug discovery and biotechnology. We discuss how data and models work together to accelerate insight, the developing infrastructure that can bring them together, and why domain experts are critical for enabling the application of artificial intelligence to scientific questions.
Generations of Artificial Intelligence: Deep Learning, Machine Learning and Statistics
AI is a term used so broadly in technical literature and popular culture, it is important for us to define what it means in the context of this article. We consider AI to be any automated system which performs tasks that would normally require human intelligence. This definition is intentionally broad, and is designed to communicate that AI is simply another tool that scientists can use to gain insights. We categorize AI approaches into three families: Statistical inference, machine learning and deep learning.
The first AI approach, direct statistical inference, is already used widely across the field. Application of, for example, T-tests or regression analysis allows scientists to determine if data compiled through a series of experiments can directly answer a critical scientific question.
Building on statistical inference, machine learning comprises methods such as logistic regression, Bayesian modeling, decision trees, random forests, and support vector machines. These employ statistical methods to find relationships between features or labels within the data, which are identified by scientists and can be detected or computed from the data. The power of machine learning is to assign statistical weights, or a measure of importance, to each feature of the data in order to create a mathematical model which can be used to gain insights from added data. The ability of scientists to combine machine learning and domain expertise to identify useful features is essential to gaining insight.
The third generation is deep learning, a powerful approach that applies machine learning methods on a massive scale. Deep learning methods capture meaning from subtle correlations that are difficult for humans to discover, and from complex higher-order interactions that are very difficult to model. Instead of putting emphasis on feature engineering, deep learning methods intrinsically identify the most informative features from underlying data. As a result, data, and as much data as possible, drawn from the greatest diversity of real conditions, and with high-quality labels, is paramount to making these methods work most effectively.
All of these generations of AI remain important tools for discovery. Each has its own strengths and weaknesses, and each has domains in which they are most useful. It is important to have a diverse toolkit to use in driving scientific discovery than to assume that a single method defines discovery. Adopting a flexible approach can pay significant dividends when it comes to maximizing the value of data found.
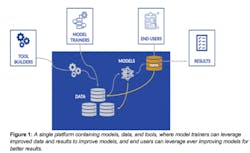
A Virtuous Cycle Between Model Trainers, Scientific Users and Data
Historically, informatics software has been separated into producers and consumers. On the producer side, academic and industry developers write tools, while on the consumer side, researchers apply these methods to their own data to drive insight. Two recent developments in AI suggest that a powerful network effect can be used to drive simultaneous development and use of AI methods.
The first development is the emergence of deep learning and machine learning frameworks (such as TensorFlow, Keras, and PyTorch) and infrastructures (platforms like DNAnexus) which allow scientists to tap into complex systems without needing to develop their own.
The second is the increasing use of large and diverse data to drive the quality of newly developed methods and, in turn, apply the models back to the data. Larger, more diverse, and higher quality datasets can be used to train better machine learning methods, which in turn can be used to generate insight.
One consequence of this is the intriguing possibility for a new type of developer-scientist: the role of a model trainer. The model trainer uses domain knowledge to identify the best, most representative datasets that capture the full scope of a problem. Then, using the frameworks that already exist, they train models that can be applied to datasets. These scientists don’t need to have all of the engineering training to actually write the frameworks and instead can directly apply their expertise to apply the frameworks in the best ways to their data. Another consequence of this is that methods can continually improve as data accumulates in the environment. The better the models, the more users are drawn to bring their own data to draw insights. By doing so, there is an opportunity to pull in that data and the insights it generates to train improved models. The models can also generate derived data and predictions that can further improve other methods. This positive feedback cycle suggests a future where methods can rapidly improve directly through their use and adoption by the community, rather than requiring long and costly software development cycles to release new and improved versions of the software.
An Ecosystem for Machine Learning in Functional Genomics — Kipoi Model Zoo
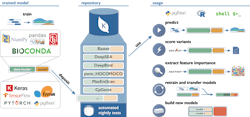
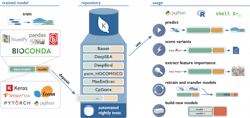
One example of this concept in action is the Kipoi Model Zoo developed in collaboration between groups at the European Bioinformatics Institute, Technical University of Munich and Stanford University. Kipoi contains an environment where researchers can upload their machine learning models with a specific focus on building models around molecular phenotype and functional genomics. The model zoo relies heavily on data from ENCODE as a base. The models in it allow researchers to predict a wide variety of applications, for example, whether a variant in a dataset disrupts a transcription factor binding site, or what the chromatin state for a region is likely to be.
Sites generating datasets similar to ENCODE (such as new CHiP-Seq, Hi-C, or ATAC-Seq datasets in different disease states, cell types or organisms) will be able to apply these data to train new models in the same way. If these models are contributed back to Kipoi or a similar framework, they will expand the methods that can quickly be applied to evaluate new data. At the same time, any researcher will be able to develop new models and methods for the data already available, ensuring that the available tools continually expand. Working with the authors of Kipoi, an execution environment was built for its models to be able to run on any data within the infrastructure platform, which allows fast iteration of this cycle as the data and models live on a single platform. In addition, these models can be applied at scale to increasingly larger datasets that live on the platform.
Image: Kipoi.org
The Power of Combining Data and Models
To demonstrate the power of having data and models on a single platform, we combined the models in Kipoi Model Zoo with some of the extensive cancer genome data on the St. Jude Pediatric Cancer Cloud. Developed by St. Jude Children’s Research Hospital in conjunction with strategic partners, St. Jude Cloud is the world’s largest public repository of pediatric cancer genomics data and offers unique analysis tools and visualizations in a secure cloud-based environment.
Starting from a set of somatic variants called across 700 tumor-normal pairs in multiple cancer types, the available 500-plus DeepBind Transcription Factor models in Kipoi were applied to predict whether variants disrupt transcription factor binding sites. This method revealed a number of transcription factors which were more or less likely to be disrupted by somatic variants when compared to match control positions nearby to the variants. Not only is this discovery is scientifically interesting, this large-scale analysis completed in less than a day, because both the data and the models are in a single scalable computing environment. Having data in standard formats, models that can read these formats, and an infrastructure that allows their rapid combination can dramatically reduce time to insight.
Furthermore, if these results motivate follow-up experiments to confirm the predictions, it may produce new RNA-Seq and CHiP-Seq data which can be used to further improve the models, leading to the next iteration of the data-model-insight cycle. With new and diverse data types available, model trainers with sufficient domain expertise can rapidly improve deep learning and machine learning models by retraining models on new, biologically relevant data.
Unlocking the Importance of Domain Expertise — Collaborations to Improve DeepVariant
Deep learning methods are powerful because they are able to discover features within the data that a human would not be able to consider, either because they are impossible to enumerate, don’t seem significant, or can’t be easily identified. However, to do this, the deep learning methods must be able to see data which represents as great a diversity of the problem as possible. For this, model trainers must have sufficient domain expertise to be able to identify diverse data types that are relevant to their field.
Collaborations with GoogleBrain around DeepVariant have demonstrated how domain expertise drives improvement to deep learning models. Google developed DeepVariant as a deep learning-based variant caller which performed with the highest overall accuracy in the PrecisionFDA Truth Challenge, beating out other industry recognized methods such as GATK. After winning the competition, Google wanted to see if DeepVariant could be further assessed and improved by identifying diverse datasets. It was subsequently found that DeepVariant and most other variant callers perform poorly on sequencing preparation that uses PCR. When DeepVariant was trained, it had only been trained on PCR-Free sequencing preparations.
One powerful principle of deep learning is that once a realization like this is made, the method can be improved without needing to write a single additional line of code. By simply providing training genomes, which had been prepared with PCR, the DeepVariant model is able to learn the error profiles characteristic in PCR-prepared data.
The key to driving improvements in deep learning models is the domain knowledge about genomics. An expert had to recognize that data representing PCR library preparation might not have been captured in the training datasets considered, and by adding PCR data to the training sets could provide deep learning models new insight to the problem. The deep learning approaches may be able to quickly learn from the data, but an expert is needed to find what should go into their curriculum. Rather than replacing domain experts as a black box application, deep learning has the potential to become a powerful tool for domain experts to drive further insights. As more data is generated, the ability to collaborate between domain experts and machine learning experts will be paramount.
Conclusion
From statistical inference to deep learning, artificial intelligence is a powerful set of tools for scientists to use to drive discovery. However, artificial intelligence is no magic bullet — it’s reliant on the data that is available and, as more data becomes available for the development of new methods, domain expertise will be critical for knowing which data and methods to use for the best results. In turn, domain experts turned model trainers will be able to rapidly improve methods by supplying larger and more diverse data, creating a self-driving loop between models and data to generate insight. Fully harnessing that power will require establishing an environment where data, infrastructure, and expertise can seamlessly combine in as streamlined a manner as possible. Such a framework can significantly accelerate both method development and scientific discovery, as domain experts and machine learning experts collaborate together on driving the data-model-insight loop.