Managing Objectionable Events in cGMP Cleanrooms: A Polyphasic Approach
In biopharmaceutical manufacturing, an accurate and comprehensive knowledge of the entire process flow is a critical part of a thorough cGMP microbial control program. The goal is to have precise data highlighting where incursion points may exist, and control those points to limit or prevent migration into the process flow. Historically, the identification of cleanroom environmental monitoring (EM) isolates has been limited to the genus or group level for most organisms, with some ability to provide species level identification only if absolutely required.
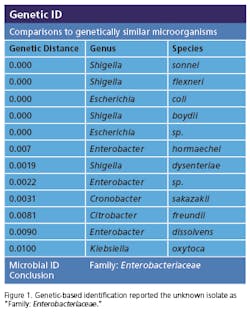
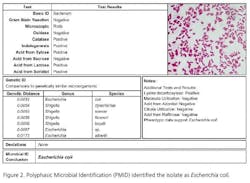
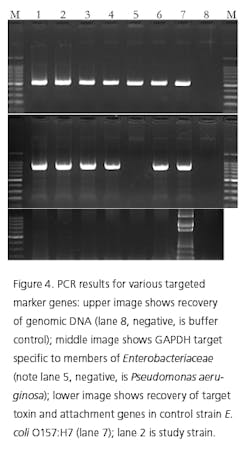
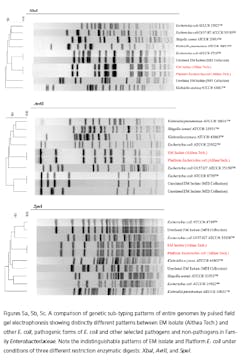
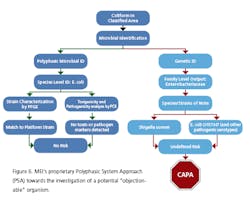
Recently, rapid technologies have become available, especially DNA-sequencing based systems, to more quickly identify microorganisms. These techniques can help manufacturers to understand and investigate potential physical and temporal sources of contamination. However, while such genetic-based systems may be rapid, they are still inherently limited by their inability to discriminate between species and some genera in some critical categories. Additionally, an identification match is only possible if the organism and its reference are adequately differentiated and in the manufacturers’ database. Thus, with this increase in technology comes the likelihood of identifying a far greater number of potentially “objectionable” environmental organisms (e.g., coliforms, pathogens) than ever before, as well as challenges in knowing what to do with the additional data.It’s important, therefore, that new tools and the data they produce not take precedence over the practical application of good microbiological practices. If anything, these technologies increase the necessity for manufacturers to take more rational and manageable approaches to environmental monitoring.We recommend a polyphasic approach, one that makes use of key characteristics or properties of a particular unknown organism in tandem with its genetic information. Such an approach provides a more definitive taxonomic identification; it also avoids the pitfalls of potentially misidentifying organisms due to the limitations of various commercial phenotypic and genotypic microbial ID systems and their databases. This article will detail such an approach taken recently during work at facilities of Althea Technologies in San Diego. An Organism of ConcernIn the current study, Molecular Epidemiology Inc. (MEI) used a polyphasic identification approach, combining a genetic-based microbial ID assay (16S rRNA sequencing) with a broad spectrum for phenotypic and biochemical analyses, to accurately identify a potentially objectionable environmental organism submitted by Althea. The organism of concern was recovered during routine environmental monitoring, while aliquoting in a tissue culture hood. The procedure involved a biofermentation Master Cell Bank (MCB), with Escherichia coli as the platform.Since the polyphasic determination presented the EM isolate as belonging to the same genus and species as the MCB and was a potentially objectionable organism (coliform), further analysis of its potential for pathogenicity was required. The process included targeted PCR analysis of toxins as well as attachment factors associated with recognized pathogenic forms, such as Enterotoxigenic E. coli (ETEC), Shiga-toxin producing E. coli (STEC), Enterohemorrhagic E. coli (EHEC), and Enteropathogenic E. coli (EPEC), or closely related species such as Shigella spp.The following organisms were used for Quality Control purposes in the various analytical tests performed: E. coli ATCC 8739, Shigella sonnei ATCC 25931, Klebsiella pneumoniae ATCC 10031, and Klebsiella oxytoca ATCC 43863. These were obtained as lyophilized cultures, reconstituted and sub-cultured as recommended for isolated colonies. The study was further supplemented with ATCC-derived strains, Enterobacter cloacae ATCC 23355, Pseudomonas aeruginosa ATCC 9027, E. coli ATCC 25922 and E. coli O157:H7 ATCC 35150, from MEI’s QC collection as well as two additional E. coli strains from lab and environmental sources.Additionally, Althea submitted an isolate of the Master Cell Bank platform strain used in processing. Representative cultures of each microorganism were subjected to 16S rRNA sequencing, colonial, morphological, biochemical (Vitek GNI, bioMérieux) observations and DNA fingerprinting using pulsed field gel electrophoresis (PFGE analysis—with restriction enzymes XbaI, AvrII and SpeI). Analysis of toxigenic and pathogenic potential was conducted using a proprietary polymerase chain reaction (PCR) method (MEI, 2010) to determine the presence or absence of toxin markers and pathogenicity factors in the reference and subject strains.Confirming IdentificationFurther comparisons with related strains with pathogenic potential differentiated and confirmed this identification (data not shown). Of concern was the equally identical taxonomic ID presented by the QC culture of E. coli O157:H7 ATCC 35150. Furthermore, sequence alignment data (Figure 3) indicate the indistinguishable similarity in the 16S rRNA gene sequence associated with the pathogenic species. Therefore, reliance on genetic-based ID or even a microbial ID conclusion would not rule out the possibility of a frank pathogen.
The key is to understand the process and potential incursion points, and to complement them with Corrective Action/Preventive Action, based on the potential toxigenicity or pathogenicity of the organism. This, coupled with a science-based Risk Assessment, allows for a more rigorous evaluation of potential EM excursions.
References
1. Clinical Laboratory Standards Institute. Interpretive Criteria for Identification of Bacteria and Fungi by DNA Target Sequencing: Approved Guideline. CLSI document MM18-A. Wayne, PA, 2008.
2. Molecular Epidemiology, Inc. dba IEH Laboratories & Consulting Group. IEH E. coli O157, Stx-producing E. coli, (STEC) with Intiman and Salmonella Test System. AOAC-RI PTM 100701, AOAC Research Institute, Gaithersburg, MD, USA, 2010.
About the Authors
Jaspreet S. Sidhu, Ph.D. (VP, Business Development and Pharmaceutical Microbiology), Connor Tyler (Scientist), Greg Ma (Director of General Microbiology), and Mansour Samadpour, Ph.D. (President, CEO) represent Molecular Epidemiology, Inc. in Lake Forest Park, Washington. E.J. Brandreth is VP of Quality and Regulatory Affairs for Althea Technologies, San Diego.